But I was pleasantly surprised. Although Ramakrishnan had accomplished his Nobel Prize winning work by X-ray crystallography, surprisingly, he mostly talked about his recent work using cryo-electron microscopy. It was probably wrong to categorize him as an X-ray crystallographer. He is first and foremost fascinated by the ribosome - his enthusiasm was palpable - and he is willing to try anything that will help him study the ribosome. He didn't start his career doing X-ray crystallography, either. That was also just a technique that he picked up in order to study the ribosome.
Why cryo-EM? Cryo-EM has some advantages over X-ray crystallography. Above all, you don't need to crystallize your sample. The amount of material required for cryo-EM is much less than what is required for X-ray crystallography. Unlike X-ray crystallography, you can work with heterogeneous samples and even solve structures of the molecules in different conformations. However, those advantages would be of little help if the resolution of the structure you get is not high enough to give you useful information. In fact, my impression of cryo-EM structures was that of blob-like pictures that do not reveal much details.
What I didn't know, as I'm not a structural biologist and hadn't been paying enough attention, was that the resolution that can be achieved by cryo-EM has improved dramatically in recent years. According to Ramakrishnan, this improvement in resolution is due to a few key developments. One is development of better detectors. Other developments concern how the data are processed to solve the structure and they include introduction of Bayesian statistics and correcting for the movements of the molecules induced by the electron beam and so on. These developments have been documented in several recent reviews. (For example, [1], [2], and [3]. Notice that words like "revolution" and "new era" are used in the titles. Also see a couple of very recent articles[4,5].)
These advances have made it possible to obtain 3D structures of biological macromolecules in 3-5 Å range. For example, this is the structure of the yeast mitochondrial large ribosome subunit solved to the resolution of 3.2 Å by Ramakrishnan's group in collaboration with Sjors Scheres' group[6].
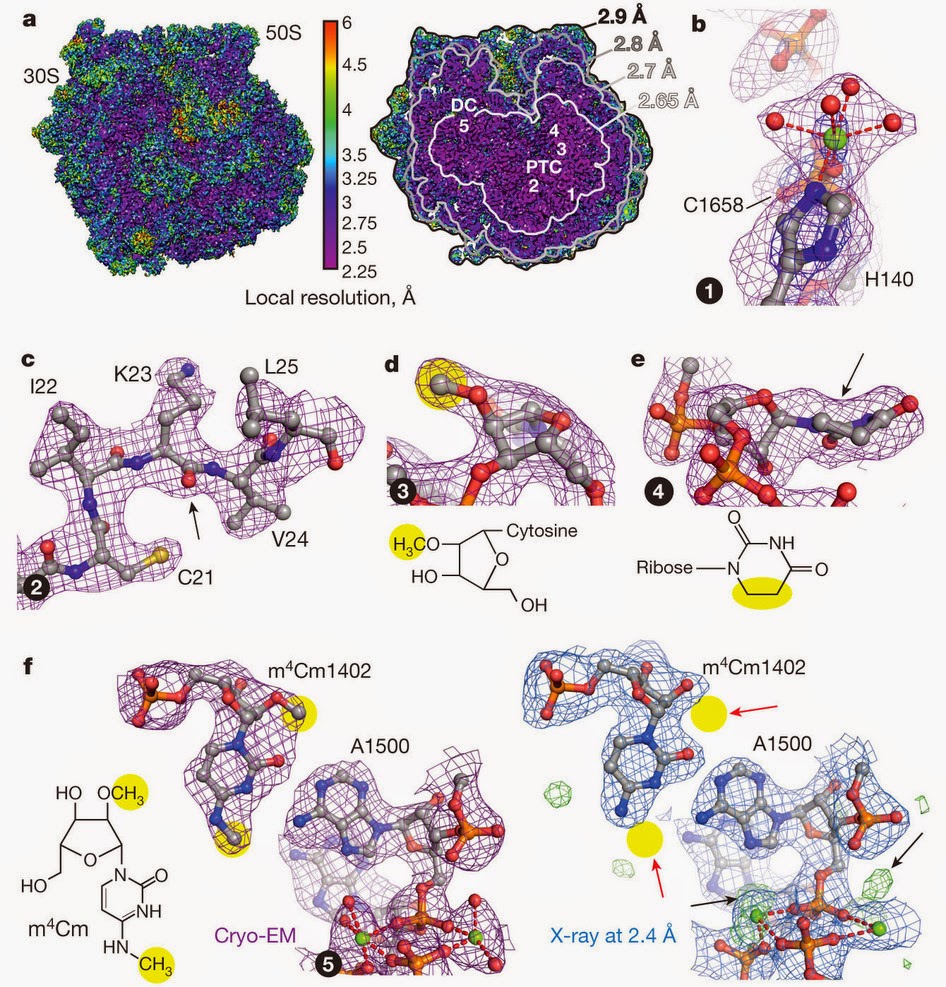
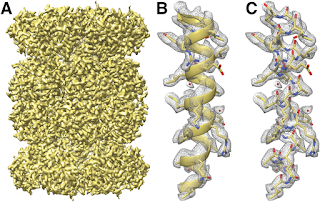
Since I attended Ramakrishnan's seminar, there have been a number of papers by various groups which reported high resolution structures using cryo-electron microscopy. For example, the following figure is the structure of E. coli ribosome by Fischer et al. that achieved the resolution of less than 3 Å[7].
The following is the structure of the human ribosome by Khatter et al.[8] with an average resolution of 3.6 Å, reaching 2.9 Å resolution at some places.
It's not just the ribosome. The following is the structure of 20S proteasome by Campbell et al. that was solved to 2.8 Å[9].
The following is the structure of anthrax protective antigen by Jiang et al. that was solved to 2.9 Å[10].
And then came this week the structure of β-galactosidase that achieved a resolution of 2.2 Å[11].
With this kind of resolution, you are able to see a very detailed structure.
I am ignorant about this field, but it seems that there have been tremendous improvements in the technique. This can have a huge impact on understanding the mechanisms of many biological macromolecules.
References
- Kühlbrandt, W. Biochemistry. The resolution revolution. Science 343, 1443-1444 (2014). [Pubmed] [Article]
- Kühlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014). [Pubmed] [Article]
- Bai, X. C., McMullan, G., & Scheres, S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 40, 49-57 (2015). [Pubmed] [Article]
- Cheng, Y., Grigorieff, N., Penczek, & P. A., Walz, T. A Primer to Single-Particle Cryo-Electron Microscopy. Cell 161, 438-449 (2015). [Pubmed] [Article]
- Cheng, Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 161, 450-457 (2015). [Pubmed] [Article]
- Amunts, A., Brown, A., Bai, X. C., Llácer, J. L., Hussain, T., Emsley, P., Long, F., Murshudov, G., Scheres, S. H., & Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485-1489 (2014). [Pubmed] [Article]
- Fischer, N., Neumann, P., Konevega, A. L., Bock, L. V., Ficner, R., Rodnina, M. V., & Stark, H. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520, 567-570 (2015). [Pubmed] [Article]
- Khatter, H., Myasnikov, A. G., Natchiar, S. K., & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015). [Pubmed] [Article]
- Campbell, M. G., Veesler, D., Cheng, A., Potter, C. S., & Carragher, B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife (2015). [Pubmed] [Article]
- Jiang, J., Pentelute, B. L., Collier, R. J., & Zhou, Z. H. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature (2015) [Epub ahead of print]. [Pubmed] [Article]
- Bartesaghi, A., Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L., & Subramaniam S. Science (2015) [Epub ahead of print]. [Pubmed] [Article]




No comments:
Post a Comment