2015 Nobel Prize in Chemistry was awarded to Tomas Lindahl, Paul Modrich, and Aziz Sancar for their studies of DNA repair. This was a huge surprise to me. There is no question that DNA repair is important. But it wasn't clear to me who should win the Nobel Prize for DNA repair. This year's Lasker Award also recognized works on DNA repair, but it went instead for Stephen Elledge and Evelyn Witkin. That selection was not obvious to me, either, although I knew that Stephen Elledge had done impressive works and had also received Gairdner Award, Rosenstiel Award, and Dickson Prize. It seemed to me that there are so many aspects to DNA repair pathways and there are many other people who have also contributed to this field. Larry Moran mentions Philip Hanawalt as someone who may have been overlooked. A commentator in In The Pipeline blog mentions Richard Kolodner and Thomas Kunkel as some other important contributors in the field while noting that Paul Modrich often beat Richard Kolodner in tight races.
This is probably a case where there is no perfect answer, but at least there seems to be a logic to the decision by the Nobel committee. The works of these three elucidated details of three DNA repair mechanisms, namely base excision repair, nucleotide excision repair, and mismatch repair. They did this by establishing the enzymatic reactions in the test tubes. So, the nature of their studies fits with the Chemistry Prize. All of these mechanisms deal with DNA damages that occur on one strand of DNA. In comparison, my impression is that someone like Stephen Elledge has worked on cellular responses when the DNA is damaged and the nature of the damages include cuts on two strands of DNA. He has mainly used genetic approaches to understand the signaling cascades and the systematic responses rather than the detailed biochemistry of the repair processes. So, what Elledge did can be considered more genetic and biological and less chemical. And of course by focusing on base excision repair, nucleotide excision repair, and mismatch repair, the Nobel committee was able to keep the number of recipients to three.
Ultimately, the Nobel committee has the right to choose the recipients that they like. We tend to scrutinize the Nobel Prize much more than we do other prizes, but the Nobel Prize is not fundamentally different from other prizes. In the past, the Nobel Prize often went to people who had already been recognized by other major prizes, but it is interesting that they made a little surprising choice this year. And considering the importance of DNA repair, they didn't make a bad choice.
Sunday, October 11, 2015
2015 Nobel Prize in Physics for neutrino oscillations
The 2015 Nobel Prize in Physics was awarded to Takaaki Kajita and Arthur McDonald for the discovery of neutrino oscillations. The existence of neutrino oscillations means that neutrinos have non-zero masses. This is a piece that does not quite fit to the original Standard Model, which has been otherwise very successful. (At least that is the extent of my knowledge.) It was one of the biggest discoveries in elementary particle physics in the last few decades before the discovery of Higgs boson and it is not a huge surprise that they finally decided to award the Physics Prize for the discovery of neutrino oscillations this year. Takaaki Kajita worked for the Super Kamiokande project in Japan. Arthur McDonald worked for the SNO project in Canada.
It was also a somewhat bittersweet announcement. Yoji Totsuka, who lead the Super Kamiokande project, is no longer alive. He passed away in 2008 at the age of 66. He most likely would have shared the Nobel Prize had he been alive.
In my previous post, I guessed that the discovery of neutrino oscillations could win the Nobel Prize in Physics. I thought this was an unquestionably important discovery. The discovery of exoplanets, predicted to win the Physics Prize by some, may be intriguing and is no doubt a great technical feat, but we already knew that planets exist, at least in our solar system. The discovery of neutrino oscillations was more fundamental and surprising. I also think that the discovery of neutrino oscillations was probably on a more solid foundation than some other contenders. The tests of Bell's inequality are important, but there are arguments of possible loopholes. The existence of dark matter was only inferred from indirect methods and we still don't know what dark matter really is. The only question was how to deal with the death of Totsuka. It seems that the answer was to choose Kajita as one of the recipients.
I'm not sure if only awarding the leaders of these big projects a prize is a good thing. Nevertheless, I'm sure that this is an exciting news for the scientists who worked on the Super Kamiokande project and the SNO project. I would like to congratulate them for the job well done.
It was also a somewhat bittersweet announcement. Yoji Totsuka, who lead the Super Kamiokande project, is no longer alive. He passed away in 2008 at the age of 66. He most likely would have shared the Nobel Prize had he been alive.
In my previous post, I guessed that the discovery of neutrino oscillations could win the Nobel Prize in Physics. I thought this was an unquestionably important discovery. The discovery of exoplanets, predicted to win the Physics Prize by some, may be intriguing and is no doubt a great technical feat, but we already knew that planets exist, at least in our solar system. The discovery of neutrino oscillations was more fundamental and surprising. I also think that the discovery of neutrino oscillations was probably on a more solid foundation than some other contenders. The tests of Bell's inequality are important, but there are arguments of possible loopholes. The existence of dark matter was only inferred from indirect methods and we still don't know what dark matter really is. The only question was how to deal with the death of Totsuka. It seems that the answer was to choose Kajita as one of the recipients.
I'm not sure if only awarding the leaders of these big projects a prize is a good thing. Nevertheless, I'm sure that this is an exciting news for the scientists who worked on the Super Kamiokande project and the SNO project. I would like to congratulate them for the job well done.
Monday, October 5, 2015
2015 Nobel Prize in Physiology or Medicine to Campbell, Omura, and Tu
This year's Nobel Prize in Physiology or Medicine was awarded to William Campbell, Satoshi Omura, and Youyou Tu. These were not the names that were included in my previous post, but, when I wrote "Something more clinical is always a possibility," this was the kind of thing that I had in my mind. Campbell and Omura are credited for discovering a drug against roundworm parasites. Tu is credited for discovering a drug against Malaria. I did know that Tu had received the Lasker Award.
I don't have any interesting thing to say because I don't know much about the works of these researchers — I tend to be more interested in discoveries in basic science because I am a basic scientist myself. However, I don't think there is any question that these researchers have made huge contributions by discovering drugs that have saved so many people. This is after all a medicine prize. I would like to congratulate them and thank them for what they accomplished.
I don't have any interesting thing to say because I don't know much about the works of these researchers — I tend to be more interested in discoveries in basic science because I am a basic scientist myself. However, I don't think there is any question that these researchers have made huge contributions by discovering drugs that have saved so many people. This is after all a medicine prize. I would like to congratulate them and thank them for what they accomplished.
Sunday, October 4, 2015
Who will win the Nobel Prize?
I have hardly written anything on this blog. The "Nobel Week" will start tomorrow. I might as well write about my speculations of who might win.
Guessing who will win the Nobel Prize has been a guilty pleasure to me. I feel a little guilty because I know it is silly — the significance of a scientific work should not depend on whether it is recognized by the Nobel Prize. Winners of the Nobel Prize are decided by humans and they are constrained by the rules. Two rules have huge influence on the choice of the winners: the Prize is awarded to a maximum of only three winners in a given category in a given year; the Prize is not awarded posthumously. In some ways, these constraints make it a little more interesting game to guess the winners. There could be some important work that deserves a recognition, but is difficult for the Nobel Committee to choose the winners for one reason or another. (I will write about some of the examples that I have in my mind far below.)
Coming up with a list of possible scientific works and scientists that have potential to win the Nobel Prize is not too difficult. There are other awards which give good indications of possible candidates. Thomson Reuters ScienceWatch has a list of people that they have predicted to win the Nobel Prize. There are many people who post their predictions on blogs and other forums. The difficult part is guessing who is more likely to win. Guessing has also become harder because scientific works that I considered to be locks have received the Nobel Prize already. Those include vesicle traffic (Medicine 2013), iPS cells (Medicine 2012), telomere (Medicine 2009), RNAi (Medicine 2006), ribosome structure (Chemistry 2009), Higgs boson (Physics 2013), spontaneous symmetry breaking and CP violation (Physics 2008), and cosmic microwave background (Physics 2006).
Below, I will try to write my thoughts on possible scientific works and scientists who might win the Nobel Prize. I will start with some guesses followed by more lengthy rundown of the topics. Because it is more meaningful to write about topics and people that I know something about, I will put more emphasis on them than more probable topics that I'm less familiar with.
Things that I think have high likelihood of winning soon
Physiology or Medicine
Protein chaperone (Arthur Horwich and F. Ulrich Hartl) or optogenetics (Gero Miesenböck, Karl Deisseroth, and Georg Nagel?). Protein chaperone work also has a chance of winning the Chemistry Prize.
Chemistry
It's likely to be something/someone I'm not familiar with, but lithium-ion batteries mentioned by many people sound very plausible.
Physics
Is this supposed to be a year for astrophysics/cosmology? People are talking about dark matter and exoplanets, but I'm not so sure. You can find a list of possible subjects far below.
Things that I want to be recognized (You can see my bias.)
Physiology or Medicine
Nuclear receptor (Pierre Chambon and Ronald Evans) AND eukaryotic transcription machineries (Robert Roeder; Chambon also worked in this area).
Chemistry
Chemical biology (Stuart Schreiber) and histone modifications (David Allis)
Physics
Neutrino oscillation (Arthur McDonald, Takaaki Kajita, Yoichiro Suzuki, or Atsuto Suzuki) or quantum entanglement (John Caluser, Alain Aspect, and Anton Zeilinger)
A few interesting subjects that haven't been mentioned by many people
Physiology or Medicine
Paleogenetics (Svante Pääbo)
Chemistry
Cryo-electron microscopy (Richard Henderson, Joachim Frank, and Sjors Scheres?)
Rundown of the subjects
Physiology or Medicine
Genome editing using CRISPR/Cas9
Possible winners: Jennifer Doudna, Emmanuelle Charpentier, Virginijus Siksnys, Feng Zhang, George Church
I don't really think that CRISPR/Cas9 will win the Nobel Prize this year, but I wanted to write it first because it is the hottest topic. This has been a true game changer — I have used it myself for my research and I know how powerful it is. Many people are speculating about a Nobel Prize for CRISPR/Cas9 even though the key papers were only published in 2012 and 2013. It is very likely to win a Nobel Prize eventually, although I don't think it will be this year. And this could be a good example of the absurdity of choosing three or less winners.
The front runners for the possible winners could be Jennifer Doudna and Emmanuelle Charpentier. They received the Breakthrough Prize in Life Sciences, among other honors. In an elegant paper published in 2012, their team demonstrated that the Cas9 protein can be programmed to cut a DNA sequence of your choice by combining with a suitable RNA molecule. As ZFNs and TALENs had already shown, an enzyme like that could be a powerful tool for gene editing.
However, the 2012 paper by the Doudna/Charpentier team didn't actually demonstrated genome editing using the CRISPR/Cas9 system. The first papers that actually accomplished genome editing by CRISPR/Cas9 came from the labs of Feng Zhang and George Church. These papers, published online in early January of 2013, were the ones that sent the shock wave. Doudna's lab also published their genome editing result later that month, but it was a little late, not as comprehensive as the papers by Zhang and Church labs, and didn't have quite the same impact.
If the Nobel Prize could be shared by four people, the choice would be easier to make. However, this is where the magic number of three becomes important.
The question is, which of these papers was the most crucial advancement. One could argue that Doudna/Charpentier paper was more important because, once you know that CRISPR/Cas9 system can be programmed to cut DNA of your choice, it was obvious to use it for genome editing in analogy with the previous techniques using ZFNs and TALENs. On the other hand, one could make a counterargument that actually showing that it can be used for genome editing in cells is not a trivial matter. Doudna admits that her lab struggled to get genome editing in the cells to work and contacted George Church whose lab had already had success. Feng Zhang and George Church had the advantage of having worked with TALENs previously.
One thing that I found odd is that a paper by Virginijus Siksnys' group in Lithuania tends to get overlooked even though they reported the activity of Cas9 about the same time as the paper by Doudna and Charpentier (and in fact submitted a little earlier). It is as if there is a fixed narrative. Charpentier also tends to be overshadowed by Doudna, but an earlier discovery of tracrRNA by Charpentier's group was crucial, so she deserves a lot of credit for that, too.
In any case, these were just a few steps of a long line of research to understand the CRISPR/Cas system. The Nobel Prize tends to put a spotlight on a few people, but it can give a distorted picture of how science advances. Personally, I would like to thank people who were studying CRISPR before it was cool, before people realized that it can be a powerful tool, and even before it was known to be a bacterial immune system.
Anyway, who will win the Nobel Prize in the end? Many different scenarios are possible:
I might add that, from a purely technological point of view, I think Feng Zhang has had the biggest impact and he will continue to as he keeps coming up with new ideas of using the CRISPR/Cas technology. Interestingly, Feng Zhang was a graduate student of Karl Deisseroth and contributed to the development of optogenetics (see below). It is really impressive that someone who is still young has already been involved in the developments of two revolutionary methods.
On the other hand, the scientist that I'm most aspired to be like is Jennifer Doudna. She is a pure scientist more interested in understanding natural phenomena than applications. In a way, it is unfortunate that many people only know her from the CRISPR work. She has done great things even before she started working on CRISPR. I think that her ribozyme crystal structure was more significant scientific achievement in her career than her CRISPR work.
Optogenetics
Possible winners: Gero Miesenböck, Karl Deisseroth, and Georg Nagel
Optogenetics is also a hot topic and is likely to win the Nobel Prize sooner than CRISPR/Cas9. I have been to a seminar by Karl Deisseroth and found it really impressive. One question, though, is if they pick optogenetics (mainly a tool for neuroscience) this year after awarding the Physiology/Medicine Prize to neuroscience last year.
Karl Deisseroth is most likely to be among the mix. Just glancing at the history of optogenetics (which I'm admittedly not too familiar with), Gero Miesenböck (who was the first to develop the technique) and Georg Nagel (who was the first to use channelrhodopsin for optogenetics) could be the other winners.
Protein chaperones
Possible winners: Arthur Horwich and F. Ulrich Hartl
This is an example of important topics in basic molecular biology that are written in textbooks. Horwich and Hartl won the Lasker Award in 2011 and shared some other major prizes. The way their accomplishments are recognized has followed a pattern that is similar to many previous Nobel Prize winners. They seem like good candidates to win the Nobel Prize anytime soon.
Nuclear receptors
Possible winners: Pierre Chambon and Ronald Evans
Like Horwich and Hartl above, Chambon and Evans won the Lasker Award in 2004 and won some other major prizes. Nuclear receptors are unquestionably important. If I'm a tiny bit hesitant to predict the Nobel Prize for Chambon and Evans, the reason is as follows. Nuclear receptors are a class of transcription factors, which are proteins that regulate transcription. When it comes to the field of transcription regulation in eukaryotic organisms, it is hard to ignore the impact of Robert Roeder. Roeder missed out when the Chemistry Prize was awarded to Roger Kornberg in 2006. A possible justification is that Kornberg's work was more structural and more fitting to the Chemistry Prize. But I'm not sure if it is fair to award Chambon and Evans ahead of Roeder. As a compromise, I wonder if choosing the trio of Roeder, Chambon, and Evans is possible. Both Roeder and Chambon discovered that eukaryotic organisms have multiple RNA polymerases. However, it's possible that Roeder lost his chance when Kornberg was the sole winner of the Chemistry Prize in 2006.
Tumor suppressor genes
Possible winners: Maybe Alfred Knudson, Thaddeus Dryja, Robert Weinberg, David Lane, Arnold Levine, or Bert Vogelstein
The question really should be why there hasn't been a Nobel Prize awarded for the discovery of tumor suppressor genes already. As a key concept in cancer biology, its importance is unquestionable. My guess is that this is a case where it is difficult to choose three (or less) clearcut winners. Many people are saying that Robert Weinberg and Bert Vogelstein should win, but the history seems a little more complicated.
Take for instance the discovery of Rb gene, whose mutation is a cause of retinoblastoma. Its discovery was reported in a paper in 1986. The authors include Robert Weinberg, arguably the biggest name in cancer biology. And sometimes he does get the credit for the discovery of Rb. However, the paper was a product of a collaboration between Weinberg's lab and Thaddeus Dryja's lab and Weinberg downplays his own role in the discovery.
Here is how Weinberg described the collaboration in the book "Natural Obsessions" by Natalie Angier:
""I (Weinberg) assured him (Dryja) that I would never try to steal his thunder," said Weinberg. "He'd done the great bulk of work in getting the probe, and anything that came of it would be credited to him. I've already had my share of glory.""
That was why Weinberg intentionally placed his name as a middle author of a seven-author paper, rather than as the last author and the corresponding author who was most responsible for the study.
If you read the book, you get the impression that the driving force for the 1986 paper was Dryja and Stephen Friend, who was a postdoc of Weinberg's lab. Weinberg's role seems to be that of the PI of a lab that allowed the project to happen. Would it be appropriate to give Weinberg the credit of discovering Rb? Most PIs would happy to take the credit, but Weinberg is on the record of saying that he didn't contribute much. Or should Stephen Friend get the credit instead? He may have done the lion's share of the work for cloning of Rb. But he was also just one of several of Weinberg's trainees who worked on the project and he had only worked a relatively short time before the publication of the paper. Dryja probably should get a credit, but he remains relatively unknown.
There are also others whose names deserve mention. For example, there is Alfred Knudson, whose two-hit hypothesis was very important. However, since it was a hypothesis rather than a concrete discovery, it is not a slam dunk case. People who were involved in showing that TP53 (p53) is a tumor suppressor gene may deserve some credits. But TP53 alone has many names associated with it, including David Lane, Arnold Levine, and Bert Vogelstein. It just seems difficult to pick three clear winners.
It is possible that some day the Nobel Committee will pick three people for the discovery of tumor suppressor genes. They may also take Weinberg's other contributions to cancer biology into consideration. Maybe, Knudson, Weinberg, and Dryja is a possible combination. It is equally likely that they will keep avoiding making such a decision.
Then again, the question is if someone like Robert Weinberg really needs the Nobel Prize. He is already famous and influential. As he himself said, he already had his share of glory.
Paleogenetics
A possible winner: Svante Pääbo
I'm not sure if this is the kind of field that will be recognized by the Nobel Prize — I haven't seen it mentioned as a possible subject for the Nobel Prize elsewhere. But I think there have been a lot of exciting developments and the Nobel Committee may decide to think outside of the box. I'm not too familiar with this field, but Svante Pääbo is a big name.
Some other topics that could be recognized by the Physiology/Medicine Prize:
Unfolded protein response (Peter Walter and Kazutoshi Mori)
Autophagy (Yoshinori Ohsumi)
Molecular motors (Michael Sheetz, James Spudich, and Ronald Vale) — It's too bad that Hugh Huxley didn't win.
Micro RNA (Victor Ambros and Gary Ruvkun)
Sensing of pain and heat (David Julius)
Hearing (James Hudspeth and David Corey)
Circadian rhythm (Jeffrey Hall, Michael Rosbash, and Michel Young) — It's too bad that Seymour Benzer didn't win.
Something more clinical is always a possibility.
Chemistry
Disclaimer: I don't know too much about chemistry, so I will only write on topics that are related to biology. Also keep in mind that some of the topics and names that I considered for the Physiology/Medicine Prize may win the Chemistry Prize instead.
Cryo-electron microscopy
Possible winners: Richard Henderson, Joachim Frank, and Sjors Scheres
I don't think it will be this year because a lot of advances happened in recent years and I also think it is unlikely that this will be the topic right after the Chemistry Prize was awarded for super-resolution (optical) microscopy last year. However, as I have written previously, there is a revolution going on in the field of cryo-electron microscopy. You can feel the excitement from reading a recent news article in Nature. I don't know too much about the field, but Richard Henderson seems to be someone who has done very important work in the past and has been trying to push the technology. Sjors Scheres seems to be credited for a new algorithm for solving the structure. Someone like Joachim Frank could be credited as an early pioneer of single particle reconstruction. Since this revolution depended on the new detectors, someone could be credited for the development of the detectors.
Chemical biology
Possible winners: Stuart Schreiber and others
Chemical biology is somewhat a vague term, but it could be summarized as clever use of chemistry, including use of small molecules, for molecular biology. Stuart Schreiber is a name that is often mentioned, although there are others. I know a little bit about Schreiber's work on "dimerizer", rapamycin, and HDAC (see below) among others. I think he did a lot of important works, but, if he wins, it will be more for a lifetime achievement than for a specific work.
The role of histone modifications in regulation of gene expression
Possible winner: David Allis
I mentioned about transcription regulation in eukaryotic organisms when I wrote about nuclear receptors above. For a long time, big discoveries in the field mostly concerned the basal transcription machinery. But a big change happened in 1996 by publication of two papers. One of the papers was published by David Allis' lab and described a discovery of a histone acetylase. They discovered the enzyme in tetrahymena, but, importantly, the yeast homologue had been known to be an activator of transcription. And of course there are human homologues as well. The other paper was from Stuart Schreiber's lab (see the entry on chemical biology above). This paper was like a mirror image to the Allis paper in that they discovered a mammalian histone deacetylase and its yeast homologue had been known to be a repressor of transcription. These papers suddenly brought histone modifications and chromatin structure into the spotlight. It was followed by the discoveries that many genes that are mutated in cancers and in some hereditary diseases encode enzymes that modify histones.
Since Schreiber's lab is not focused on histones or transcription, they didn't do much work in this area afterwards. (They did many important things in other fields, both before and after.) Allis, on the other hand, has been a leader in the study of histones and chromatin. His work doesn't have the breadth of Schreiber, but he has had a huge impact as a specialist in this field that I found exciting. It may be more appropriate to categorize him as a Physiology/Medicine Prize candidate, but I'm wondering about the outside chance of pairing Allis with Schreiber.
Physics
Neutrino oscillations
Possible winners: Arthur McDonald, Takaaki Kajita, Yoichiro Suzuki, or Atsuto Suzuki
Before the discovery of the Higgs boson a few years ago, elementary particle physics was having a somewhat stagnant period. Most of the elementary particles in the Standard Model had already been discovered and there weren't many excitements. But neutrino physics seemed to be an exception. There were exciting developments that centered around the discovery of neutrino oscillation. The Nobel Prize in Physics was awarded for neutrino physics in 2002, but the citation seemed to indicate a room for a separate prize for the discovery of neutrino oscillation.
Why hasn't there been a Nobel Prize for neutrino oscillation already? The most likely reason I can think of is untimely death of Yoji Totsuka. The candidates for winning the Nobel Prize for neutrino oscillation were Totsuka, who lead the Super Kamiokande project, and Arthur McDonald, who lead the SNO project. But Totsuka passed away in 2008. (The thought that occurred to me after hearing Totsuka's death was that they should give the Prize to Yoichiro Nambu before it's too late. They indeed awarded Nambu the Physics Prize in 2008. Nambu passed away earlier this year at the age of 94.)
I imagine that Totsuka's death created a conundrum for the Nobel Committee. If they want to award the Nobel Prize for neutrino oscillation, what is the right thing to do? You can't award the Prize posthumously to Totsuka. Should they award McDonald alone? Sometimes some of the scientists who did an important work die and only the surviving members receive the Nobel Prize. The 2013 Physics Prize was given to Englert and Higgs, who were alive, even though Englert's work was done with Brout, who had passed away in 2011. However, the discovery of neutrino oscillation was done by huge experimental teams unlike the theoretical work of Englert, Brout, and Higgs. If they only award the Prize to McDonald, it is as if to only credit the SNO project without acknowledging the Super Kamiokande project.
I am guessing that the committee has been avoiding a decision. But they may make a decision at some point if they think that the discovery of neutrino oscillations merits the Nobel Prize. One way to solve the problem is to award the Nobel Prize to someone from the Super Kamiokande project as a replacement for Totsuka. Takaaki Kajita and Yoichiro Suzuki have been mentioned as possibilities. Atsuto Suzuki of the KamLAND project is another possibility to share the prize.
A situation like this also illustrates the absurdity of the Nobel Prize or prizes in general. If these works are done by huge teams, is it appropriate to only credit the heads of such teams?
Quantum entanglement
Possible winners: John Caluser, Alain Aspect, and Anton Zeilinger
It's too bad that John Bell passed away in 1990. But quantum entanglement is weird and is such a fundamental part of quantum mechanics that experimental tests of Bell's inequality seems important enough to merit a Nobel Prize. One negative point is that the Nobel Committee went for different winners when they awarded the Prize in 2012 for works related to quantum mechanics and measurements. Are they afraid of possible loopholes?
Some other topics that could be recognized by the Physics Prize:
Topological insulators
Some other topics related to quantum information
Discovery of exoplanets
Dark matter
Guessing who will win the Nobel Prize has been a guilty pleasure to me. I feel a little guilty because I know it is silly — the significance of a scientific work should not depend on whether it is recognized by the Nobel Prize. Winners of the Nobel Prize are decided by humans and they are constrained by the rules. Two rules have huge influence on the choice of the winners: the Prize is awarded to a maximum of only three winners in a given category in a given year; the Prize is not awarded posthumously. In some ways, these constraints make it a little more interesting game to guess the winners. There could be some important work that deserves a recognition, but is difficult for the Nobel Committee to choose the winners for one reason or another. (I will write about some of the examples that I have in my mind far below.)
Coming up with a list of possible scientific works and scientists that have potential to win the Nobel Prize is not too difficult. There are other awards which give good indications of possible candidates. Thomson Reuters ScienceWatch has a list of people that they have predicted to win the Nobel Prize. There are many people who post their predictions on blogs and other forums. The difficult part is guessing who is more likely to win. Guessing has also become harder because scientific works that I considered to be locks have received the Nobel Prize already. Those include vesicle traffic (Medicine 2013), iPS cells (Medicine 2012), telomere (Medicine 2009), RNAi (Medicine 2006), ribosome structure (Chemistry 2009), Higgs boson (Physics 2013), spontaneous symmetry breaking and CP violation (Physics 2008), and cosmic microwave background (Physics 2006).
Below, I will try to write my thoughts on possible scientific works and scientists who might win the Nobel Prize. I will start with some guesses followed by more lengthy rundown of the topics. Because it is more meaningful to write about topics and people that I know something about, I will put more emphasis on them than more probable topics that I'm less familiar with.
Things that I think have high likelihood of winning soon
Physiology or Medicine
Protein chaperone (Arthur Horwich and F. Ulrich Hartl) or optogenetics (Gero Miesenböck, Karl Deisseroth, and Georg Nagel?). Protein chaperone work also has a chance of winning the Chemistry Prize.
Chemistry
It's likely to be something/someone I'm not familiar with, but lithium-ion batteries mentioned by many people sound very plausible.
Physics
Is this supposed to be a year for astrophysics/cosmology? People are talking about dark matter and exoplanets, but I'm not so sure. You can find a list of possible subjects far below.
Things that I want to be recognized (You can see my bias.)
Physiology or Medicine
Nuclear receptor (Pierre Chambon and Ronald Evans) AND eukaryotic transcription machineries (Robert Roeder; Chambon also worked in this area).
Chemistry
Chemical biology (Stuart Schreiber) and histone modifications (David Allis)
Physics
Neutrino oscillation (Arthur McDonald, Takaaki Kajita, Yoichiro Suzuki, or Atsuto Suzuki) or quantum entanglement (John Caluser, Alain Aspect, and Anton Zeilinger)
A few interesting subjects that haven't been mentioned by many people
Physiology or Medicine
Paleogenetics (Svante Pääbo)
Chemistry
Cryo-electron microscopy (Richard Henderson, Joachim Frank, and Sjors Scheres?)
Rundown of the subjects
Physiology or Medicine
Genome editing using CRISPR/Cas9
Possible winners: Jennifer Doudna, Emmanuelle Charpentier, Virginijus Siksnys, Feng Zhang, George Church
I don't really think that CRISPR/Cas9 will win the Nobel Prize this year, but I wanted to write it first because it is the hottest topic. This has been a true game changer — I have used it myself for my research and I know how powerful it is. Many people are speculating about a Nobel Prize for CRISPR/Cas9 even though the key papers were only published in 2012 and 2013. It is very likely to win a Nobel Prize eventually, although I don't think it will be this year. And this could be a good example of the absurdity of choosing three or less winners.
The front runners for the possible winners could be Jennifer Doudna and Emmanuelle Charpentier. They received the Breakthrough Prize in Life Sciences, among other honors. In an elegant paper published in 2012, their team demonstrated that the Cas9 protein can be programmed to cut a DNA sequence of your choice by combining with a suitable RNA molecule. As ZFNs and TALENs had already shown, an enzyme like that could be a powerful tool for gene editing.
However, the 2012 paper by the Doudna/Charpentier team didn't actually demonstrated genome editing using the CRISPR/Cas9 system. The first papers that actually accomplished genome editing by CRISPR/Cas9 came from the labs of Feng Zhang and George Church. These papers, published online in early January of 2013, were the ones that sent the shock wave. Doudna's lab also published their genome editing result later that month, but it was a little late, not as comprehensive as the papers by Zhang and Church labs, and didn't have quite the same impact.
If the Nobel Prize could be shared by four people, the choice would be easier to make. However, this is where the magic number of three becomes important.
The question is, which of these papers was the most crucial advancement. One could argue that Doudna/Charpentier paper was more important because, once you know that CRISPR/Cas9 system can be programmed to cut DNA of your choice, it was obvious to use it for genome editing in analogy with the previous techniques using ZFNs and TALENs. On the other hand, one could make a counterargument that actually showing that it can be used for genome editing in cells is not a trivial matter. Doudna admits that her lab struggled to get genome editing in the cells to work and contacted George Church whose lab had already had success. Feng Zhang and George Church had the advantage of having worked with TALENs previously.
One thing that I found odd is that a paper by Virginijus Siksnys' group in Lithuania tends to get overlooked even though they reported the activity of Cas9 about the same time as the paper by Doudna and Charpentier (and in fact submitted a little earlier). It is as if there is a fixed narrative. Charpentier also tends to be overshadowed by Doudna, but an earlier discovery of tracrRNA by Charpentier's group was crucial, so she deserves a lot of credit for that, too.
In any case, these were just a few steps of a long line of research to understand the CRISPR/Cas system. The Nobel Prize tends to put a spotlight on a few people, but it can give a distorted picture of how science advances. Personally, I would like to thank people who were studying CRISPR before it was cool, before people realized that it can be a powerful tool, and even before it was known to be a bacterial immune system.
Anyway, who will win the Nobel Prize in the end? Many different scenarios are possible:
- Jennifer Doudna and Emmanuelle Charpentier for showing the activity of Cas9 in test tubes and a clever use of chimeric RNA.
- Feng Zhang and George Church for the first demonstrations of genome editing using Cas9.
- Jennifer Doudna, Feng Zhang, and George Church, where Doudna gets partial credits for the demonstration of genome editing in cells and perhaps her structural work as well.
- Jennifer Doudna, Emmanuelle Charpentier, and Feng Zhang if Church's work is considered not independent of Zhang's
- Jennifer Doudna, Emmanuelle Charpentier, and Virginijus Siksnys for showing the activity of Cas9 in the test tubes.
I might add that, from a purely technological point of view, I think Feng Zhang has had the biggest impact and he will continue to as he keeps coming up with new ideas of using the CRISPR/Cas technology. Interestingly, Feng Zhang was a graduate student of Karl Deisseroth and contributed to the development of optogenetics (see below). It is really impressive that someone who is still young has already been involved in the developments of two revolutionary methods.
On the other hand, the scientist that I'm most aspired to be like is Jennifer Doudna. She is a pure scientist more interested in understanding natural phenomena than applications. In a way, it is unfortunate that many people only know her from the CRISPR work. She has done great things even before she started working on CRISPR. I think that her ribozyme crystal structure was more significant scientific achievement in her career than her CRISPR work.
Optogenetics
Possible winners: Gero Miesenböck, Karl Deisseroth, and Georg Nagel
Optogenetics is also a hot topic and is likely to win the Nobel Prize sooner than CRISPR/Cas9. I have been to a seminar by Karl Deisseroth and found it really impressive. One question, though, is if they pick optogenetics (mainly a tool for neuroscience) this year after awarding the Physiology/Medicine Prize to neuroscience last year.
Karl Deisseroth is most likely to be among the mix. Just glancing at the history of optogenetics (which I'm admittedly not too familiar with), Gero Miesenböck (who was the first to develop the technique) and Georg Nagel (who was the first to use channelrhodopsin for optogenetics) could be the other winners.
Protein chaperones
Possible winners: Arthur Horwich and F. Ulrich Hartl
This is an example of important topics in basic molecular biology that are written in textbooks. Horwich and Hartl won the Lasker Award in 2011 and shared some other major prizes. The way their accomplishments are recognized has followed a pattern that is similar to many previous Nobel Prize winners. They seem like good candidates to win the Nobel Prize anytime soon.
Nuclear receptors
Possible winners: Pierre Chambon and Ronald Evans
Like Horwich and Hartl above, Chambon and Evans won the Lasker Award in 2004 and won some other major prizes. Nuclear receptors are unquestionably important. If I'm a tiny bit hesitant to predict the Nobel Prize for Chambon and Evans, the reason is as follows. Nuclear receptors are a class of transcription factors, which are proteins that regulate transcription. When it comes to the field of transcription regulation in eukaryotic organisms, it is hard to ignore the impact of Robert Roeder. Roeder missed out when the Chemistry Prize was awarded to Roger Kornberg in 2006. A possible justification is that Kornberg's work was more structural and more fitting to the Chemistry Prize. But I'm not sure if it is fair to award Chambon and Evans ahead of Roeder. As a compromise, I wonder if choosing the trio of Roeder, Chambon, and Evans is possible. Both Roeder and Chambon discovered that eukaryotic organisms have multiple RNA polymerases. However, it's possible that Roeder lost his chance when Kornberg was the sole winner of the Chemistry Prize in 2006.
Tumor suppressor genes
Possible winners: Maybe Alfred Knudson, Thaddeus Dryja, Robert Weinberg, David Lane, Arnold Levine, or Bert Vogelstein
The question really should be why there hasn't been a Nobel Prize awarded for the discovery of tumor suppressor genes already. As a key concept in cancer biology, its importance is unquestionable. My guess is that this is a case where it is difficult to choose three (or less) clearcut winners. Many people are saying that Robert Weinberg and Bert Vogelstein should win, but the history seems a little more complicated.
Take for instance the discovery of Rb gene, whose mutation is a cause of retinoblastoma. Its discovery was reported in a paper in 1986. The authors include Robert Weinberg, arguably the biggest name in cancer biology. And sometimes he does get the credit for the discovery of Rb. However, the paper was a product of a collaboration between Weinberg's lab and Thaddeus Dryja's lab and Weinberg downplays his own role in the discovery.
Here is how Weinberg described the collaboration in the book "Natural Obsessions" by Natalie Angier:
""I (Weinberg) assured him (Dryja) that I would never try to steal his thunder," said Weinberg. "He'd done the great bulk of work in getting the probe, and anything that came of it would be credited to him. I've already had my share of glory.""
That was why Weinberg intentionally placed his name as a middle author of a seven-author paper, rather than as the last author and the corresponding author who was most responsible for the study.
If you read the book, you get the impression that the driving force for the 1986 paper was Dryja and Stephen Friend, who was a postdoc of Weinberg's lab. Weinberg's role seems to be that of the PI of a lab that allowed the project to happen. Would it be appropriate to give Weinberg the credit of discovering Rb? Most PIs would happy to take the credit, but Weinberg is on the record of saying that he didn't contribute much. Or should Stephen Friend get the credit instead? He may have done the lion's share of the work for cloning of Rb. But he was also just one of several of Weinberg's trainees who worked on the project and he had only worked a relatively short time before the publication of the paper. Dryja probably should get a credit, but he remains relatively unknown.
There are also others whose names deserve mention. For example, there is Alfred Knudson, whose two-hit hypothesis was very important. However, since it was a hypothesis rather than a concrete discovery, it is not a slam dunk case. People who were involved in showing that TP53 (p53) is a tumor suppressor gene may deserve some credits. But TP53 alone has many names associated with it, including David Lane, Arnold Levine, and Bert Vogelstein. It just seems difficult to pick three clear winners.
It is possible that some day the Nobel Committee will pick three people for the discovery of tumor suppressor genes. They may also take Weinberg's other contributions to cancer biology into consideration. Maybe, Knudson, Weinberg, and Dryja is a possible combination. It is equally likely that they will keep avoiding making such a decision.
Then again, the question is if someone like Robert Weinberg really needs the Nobel Prize. He is already famous and influential. As he himself said, he already had his share of glory.
Paleogenetics
A possible winner: Svante Pääbo
I'm not sure if this is the kind of field that will be recognized by the Nobel Prize — I haven't seen it mentioned as a possible subject for the Nobel Prize elsewhere. But I think there have been a lot of exciting developments and the Nobel Committee may decide to think outside of the box. I'm not too familiar with this field, but Svante Pääbo is a big name.
Some other topics that could be recognized by the Physiology/Medicine Prize:
Unfolded protein response (Peter Walter and Kazutoshi Mori)
Autophagy (Yoshinori Ohsumi)
Molecular motors (Michael Sheetz, James Spudich, and Ronald Vale) — It's too bad that Hugh Huxley didn't win.
Micro RNA (Victor Ambros and Gary Ruvkun)
Sensing of pain and heat (David Julius)
Hearing (James Hudspeth and David Corey)
Circadian rhythm (Jeffrey Hall, Michael Rosbash, and Michel Young) — It's too bad that Seymour Benzer didn't win.
Something more clinical is always a possibility.
Chemistry
Disclaimer: I don't know too much about chemistry, so I will only write on topics that are related to biology. Also keep in mind that some of the topics and names that I considered for the Physiology/Medicine Prize may win the Chemistry Prize instead.
Cryo-electron microscopy
Possible winners: Richard Henderson, Joachim Frank, and Sjors Scheres
I don't think it will be this year because a lot of advances happened in recent years and I also think it is unlikely that this will be the topic right after the Chemistry Prize was awarded for super-resolution (optical) microscopy last year. However, as I have written previously, there is a revolution going on in the field of cryo-electron microscopy. You can feel the excitement from reading a recent news article in Nature. I don't know too much about the field, but Richard Henderson seems to be someone who has done very important work in the past and has been trying to push the technology. Sjors Scheres seems to be credited for a new algorithm for solving the structure. Someone like Joachim Frank could be credited as an early pioneer of single particle reconstruction. Since this revolution depended on the new detectors, someone could be credited for the development of the detectors.
Chemical biology
Possible winners: Stuart Schreiber and others
Chemical biology is somewhat a vague term, but it could be summarized as clever use of chemistry, including use of small molecules, for molecular biology. Stuart Schreiber is a name that is often mentioned, although there are others. I know a little bit about Schreiber's work on "dimerizer", rapamycin, and HDAC (see below) among others. I think he did a lot of important works, but, if he wins, it will be more for a lifetime achievement than for a specific work.
The role of histone modifications in regulation of gene expression
Possible winner: David Allis
I mentioned about transcription regulation in eukaryotic organisms when I wrote about nuclear receptors above. For a long time, big discoveries in the field mostly concerned the basal transcription machinery. But a big change happened in 1996 by publication of two papers. One of the papers was published by David Allis' lab and described a discovery of a histone acetylase. They discovered the enzyme in tetrahymena, but, importantly, the yeast homologue had been known to be an activator of transcription. And of course there are human homologues as well. The other paper was from Stuart Schreiber's lab (see the entry on chemical biology above). This paper was like a mirror image to the Allis paper in that they discovered a mammalian histone deacetylase and its yeast homologue had been known to be a repressor of transcription. These papers suddenly brought histone modifications and chromatin structure into the spotlight. It was followed by the discoveries that many genes that are mutated in cancers and in some hereditary diseases encode enzymes that modify histones.
Since Schreiber's lab is not focused on histones or transcription, they didn't do much work in this area afterwards. (They did many important things in other fields, both before and after.) Allis, on the other hand, has been a leader in the study of histones and chromatin. His work doesn't have the breadth of Schreiber, but he has had a huge impact as a specialist in this field that I found exciting. It may be more appropriate to categorize him as a Physiology/Medicine Prize candidate, but I'm wondering about the outside chance of pairing Allis with Schreiber.
Physics
Neutrino oscillations
Possible winners: Arthur McDonald, Takaaki Kajita, Yoichiro Suzuki, or Atsuto Suzuki
Before the discovery of the Higgs boson a few years ago, elementary particle physics was having a somewhat stagnant period. Most of the elementary particles in the Standard Model had already been discovered and there weren't many excitements. But neutrino physics seemed to be an exception. There were exciting developments that centered around the discovery of neutrino oscillation. The Nobel Prize in Physics was awarded for neutrino physics in 2002, but the citation seemed to indicate a room for a separate prize for the discovery of neutrino oscillation.
Why hasn't there been a Nobel Prize for neutrino oscillation already? The most likely reason I can think of is untimely death of Yoji Totsuka. The candidates for winning the Nobel Prize for neutrino oscillation were Totsuka, who lead the Super Kamiokande project, and Arthur McDonald, who lead the SNO project. But Totsuka passed away in 2008. (The thought that occurred to me after hearing Totsuka's death was that they should give the Prize to Yoichiro Nambu before it's too late. They indeed awarded Nambu the Physics Prize in 2008. Nambu passed away earlier this year at the age of 94.)
I imagine that Totsuka's death created a conundrum for the Nobel Committee. If they want to award the Nobel Prize for neutrino oscillation, what is the right thing to do? You can't award the Prize posthumously to Totsuka. Should they award McDonald alone? Sometimes some of the scientists who did an important work die and only the surviving members receive the Nobel Prize. The 2013 Physics Prize was given to Englert and Higgs, who were alive, even though Englert's work was done with Brout, who had passed away in 2011. However, the discovery of neutrino oscillation was done by huge experimental teams unlike the theoretical work of Englert, Brout, and Higgs. If they only award the Prize to McDonald, it is as if to only credit the SNO project without acknowledging the Super Kamiokande project.
I am guessing that the committee has been avoiding a decision. But they may make a decision at some point if they think that the discovery of neutrino oscillations merits the Nobel Prize. One way to solve the problem is to award the Nobel Prize to someone from the Super Kamiokande project as a replacement for Totsuka. Takaaki Kajita and Yoichiro Suzuki have been mentioned as possibilities. Atsuto Suzuki of the KamLAND project is another possibility to share the prize.
A situation like this also illustrates the absurdity of the Nobel Prize or prizes in general. If these works are done by huge teams, is it appropriate to only credit the heads of such teams?
Quantum entanglement
Possible winners: John Caluser, Alain Aspect, and Anton Zeilinger
It's too bad that John Bell passed away in 1990. But quantum entanglement is weird and is such a fundamental part of quantum mechanics that experimental tests of Bell's inequality seems important enough to merit a Nobel Prize. One negative point is that the Nobel Committee went for different winners when they awarded the Prize in 2012 for works related to quantum mechanics and measurements. Are they afraid of possible loopholes?
Some other topics that could be recognized by the Physics Prize:
Topological insulators
Some other topics related to quantum information
Discovery of exoplanets
Dark matter
Saturday, May 9, 2015
クライオ電子顕微鏡技術の目覚ましい進歩
何ヶ月か前に、ヴェンキ・ラマクリシュナンのセミナーに出席する機会があった。ラマクリシュナンはリボソームの構造を解明した業績でノーベル賞を取った人だ。ノーベル賞を取るような科学者を生で見るのには感慨があるけれど、正直なところ、セミナーの内容はそれほど期待していたわけではなかった。彼の研究対象は僕自身の興味からは距離がある。細胞の中でタンパク質の工場の役割をするリボソームが重要なのは言うまでもないけれど、専門家でない人間に取ってリボソームの構造をいくつも見せられても得る物があるか疑問だった。それに彼のようにすでに功成り名を遂げた人物なら、その成功に満足して新しい成果を上げなくなっていてもおかしくはない。
予想は良い意味で裏切られた。ラマクリシュナンのノーベル賞の対象になった業績はX線結晶構造解析の手法で得られた物だけれど、驚いた事に、セミナーではクライオ電子顕微鏡を使った最近の研究の話をした。そもそも彼の事をX線結晶構造解析の専門家に分類するのが間違いなのだろう。彼はまず第一にリボソームに魅惑されていて―彼の熱意は言葉の端々に感じられた―リボソームの研究に役立つ手法ならなんでも取り入れるのだろう。彼は研究生活の最初からX線結晶構造解析をしていたわけではなかった。X線結晶構造解析もリボソームの研究のために学んだ手法に過ぎない。
なぜクライオ電子顕微鏡か。クライオ電子顕微鏡はX線結晶構造解析と比べて有利な点がいくつかある。まず第一に、サンプルを結晶化する必要がない。必要なサンプルの量も結晶化するために必要な量から比べればずっと少ない。X線結晶構造解析と違って、サンプルが均一でなくても研究できるし、いくつかの違ったコンフォメーションを解析できる可能性もある。とは言え、それで得られる構造が、有益な情報を得るのに十分な解像度がなければ、そういう利点も強みにはならない。実際、僕にとって、クライオ電子顕微鏡によって得られる構造というのは、細部がよくわからない、ぶよぶよとした塊のような物という印象があった。
僕には構造生物学は専門外で、この分野の進展に注意を払っていなかったので知らなかったのだけれど、クライオ電子顕微鏡によって達成できる解像度は近年著しく改善されてきたそうだ。ラマクリシュナンによると、この解像度の進歩はいくつかの技術的な発展によるものだそうだ。一つは、すぐれた検出器が開発されたこと。その他は、データを処理して構造を再構成するための手法の進展で、具体的には、ベイズ統計の導入や、電子ビームによって分子が動くことの影響を補正する事などだ。これらの進展は、最近のいくつかの概説にまとめられている。(例えば、 [1]、[2]、[3]。タイトルに「革命」とか「新時代」といった言葉が使われている事に注目。さらに、もっと最近の概説も参照[4,5]。)
こういった進展によって、3-5オングストロム程度の解像度の生体分子の3次元構造を得る事が可能になった。例えば、以下に示すのはラマクリシュナンのグループがSjors Scheresのグループとの共同研究で得た酵母のミトコンドリアのリボソームの大サブユニットの構造[6]。
ラマクリシュナンのセミナーの後も、クライオ電子顕微鏡を使った高解像度の構造の論文をいろいろなグループが次々に発表している。例えば、次の図はFischerらによって発表された大腸菌のリボソームの構造で、解像度は3オングストローム未満を達成している[7]。
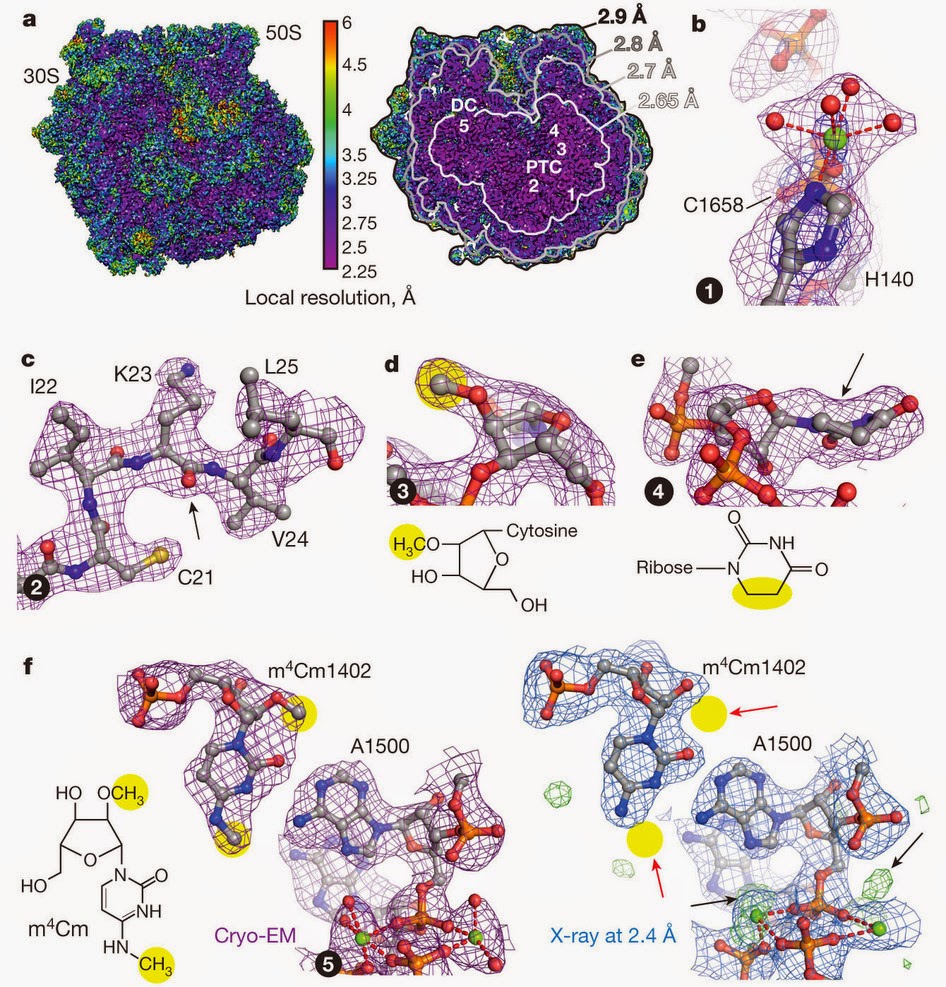
次の図はKhatterらによるヒトのリボソームの構造で[8]、解像度は平均で3.6オングストローム、部分によっては2.9オングストロームだ。
リボソームばかりではない。次の図はCampbellらによる20Sプロテアソームの構造で[9]、解像度は2.8オングストローム。
次の図は、Jiangらによる炭疽菌の防御抗原の構造で[10]、解像度は2.9オングストロームだ。
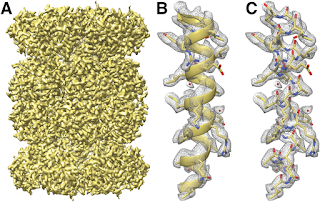
そして今週には解像度が2.2オングストロームのβガラクトシダーゼの構造が発表された[11]。
ここまで解像度が高いと、かなり細かな構造まで見える。
この分野については無知だけれど、ものすごい進歩が起きたみたいだ。生体分子のメカニズムの理解に大きなインパクトがありそうだ。
参考文献
- Kühlbrandt, W. Biochemistry. The resolution revolution. Science 343, 1443-1444 (2014). [Pubmed] [Article]
- Kühlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014). [Pubmed] [Article]
- Bai, X. C., McMullan, G., & Scheres, S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 40, 49-57 (2015). [Pubmed] [Article]
- Cheng, Y., Grigorieff, N., Penczek, & P. A., Walz, T. A Primer to Single-Particle Cryo-Electron Microscopy. Cell 161, 438-449 (2015). [Pubmed] [Article]
- Cheng, Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 161, 450-457 (2015). [Pubmed] [Article]
- Amunts, A., Brown, A., Bai, X. C., Llácer, J. L., Hussain, T., Emsley, P., Long, F., Murshudov, G., Scheres, S. H., & Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485-1489 (2014). [Pubmed] [Article]
- Fischer, N., Neumann, P., Konevega, A. L., Bock, L. V., Ficner, R., Rodnina, M. V., & Stark, H. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520, 567-570 (2015). [Pubmed] [Article]
- Khatter, H., Myasnikov, A. G., Natchiar, S. K., & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015). [Pubmed] [Article]
- Campbell, M. G., Veesler, D., Cheng, A., Potter, C. S., & Carragher, B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife (2015). [Pubmed] [Article]
- Jiang, J., Pentelute, B. L., Collier, R. J., & Zhou, Z. H. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature (2015) [Epub ahead of print]. [Pubmed] [Article]
- Bartesaghi, A., Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L., & Subramaniam S. Science (2015) [Epub ahead of print]. [Pubmed] [Article]
Remarkable Progress in Cryo-Electron Microscopy
Several months ago, I had a chance to attend a seminar by Venki Ramakrishnan. Ramakrishnan is a scientist who won a Nobel Prize for his accomplishment of solving the structure of the ribosome. While it is cool to watch a Nobel Prize winner talk, to be honest, I wasn't really expecting to get much out of the seminar. His research topic - structure of the ribosome - is far from my own interest. Ribosome, which functions as a protein factory in the cell, is undoubtedly important. But if you are not an aficionado, what can you learn from structure after structure of ribosomes? Also, for someone with his accomplishment, it is not uncommon to rest on his laurel and stop doing something new.
But I was pleasantly surprised. Although Ramakrishnan had accomplished his Nobel Prize winning work by X-ray crystallography, surprisingly, he mostly talked about his recent work using cryo-electron microscopy. It was probably wrong to categorize him as an X-ray crystallographer. He is first and foremost fascinated by the ribosome - his enthusiasm was palpable - and he is willing to try anything that will help him study the ribosome. He didn't start his career doing X-ray crystallography, either. That was also just a technique that he picked up in order to study the ribosome.
Why cryo-EM? Cryo-EM has some advantages over X-ray crystallography. Above all, you don't need to crystallize your sample. The amount of material required for cryo-EM is much less than what is required for X-ray crystallography. Unlike X-ray crystallography, you can work with heterogeneous samples and even solve structures of the molecules in different conformations. However, those advantages would be of little help if the resolution of the structure you get is not high enough to give you useful information. In fact, my impression of cryo-EM structures was that of blob-like pictures that do not reveal much details.
What I didn't know, as I'm not a structural biologist and hadn't been paying enough attention, was that the resolution that can be achieved by cryo-EM has improved dramatically in recent years. According to Ramakrishnan, this improvement in resolution is due to a few key developments. One is development of better detectors. Other developments concern how the data are processed to solve the structure and they include introduction of Bayesian statistics and correcting for the movements of the molecules induced by the electron beam and so on. These developments have been documented in several recent reviews. (For example, [1], [2], and [3]. Notice that words like "revolution" and "new era" are used in the titles. Also see a couple of very recent articles[4,5].)
These advances have made it possible to obtain 3D structures of biological macromolecules in 3-5 Å range. For example, this is the structure of the yeast mitochondrial large ribosome subunit solved to the resolution of 3.2 Å by Ramakrishnan's group in collaboration with Sjors Scheres' group[6].
Since I attended Ramakrishnan's seminar, there have been a number of papers by various groups which reported high resolution structures using cryo-electron microscopy. For example, the following figure is the structure of E. coli ribosome by Fischer et al. that achieved the resolution of less than 3 Å[7].
The following is the structure of the human ribosome by Khatter et al.[8] with an average resolution of 3.6 Å, reaching 2.9 Å resolution at some places.
It's not just the ribosome. The following is the structure of 20S proteasome by Campbell et al. that was solved to 2.8 Å[9].
The following is the structure of anthrax protective antigen by Jiang et al. that was solved to 2.9 Å[10].
And then came this week the structure of β-galactosidase that achieved a resolution of 2.2 Å[11].
With this kind of resolution, you are able to see a very detailed structure.
I am ignorant about this field, but it seems that there have been tremendous improvements in the technique. This can have a huge impact on understanding the mechanisms of many biological macromolecules.
References
But I was pleasantly surprised. Although Ramakrishnan had accomplished his Nobel Prize winning work by X-ray crystallography, surprisingly, he mostly talked about his recent work using cryo-electron microscopy. It was probably wrong to categorize him as an X-ray crystallographer. He is first and foremost fascinated by the ribosome - his enthusiasm was palpable - and he is willing to try anything that will help him study the ribosome. He didn't start his career doing X-ray crystallography, either. That was also just a technique that he picked up in order to study the ribosome.
Why cryo-EM? Cryo-EM has some advantages over X-ray crystallography. Above all, you don't need to crystallize your sample. The amount of material required for cryo-EM is much less than what is required for X-ray crystallography. Unlike X-ray crystallography, you can work with heterogeneous samples and even solve structures of the molecules in different conformations. However, those advantages would be of little help if the resolution of the structure you get is not high enough to give you useful information. In fact, my impression of cryo-EM structures was that of blob-like pictures that do not reveal much details.
What I didn't know, as I'm not a structural biologist and hadn't been paying enough attention, was that the resolution that can be achieved by cryo-EM has improved dramatically in recent years. According to Ramakrishnan, this improvement in resolution is due to a few key developments. One is development of better detectors. Other developments concern how the data are processed to solve the structure and they include introduction of Bayesian statistics and correcting for the movements of the molecules induced by the electron beam and so on. These developments have been documented in several recent reviews. (For example, [1], [2], and [3]. Notice that words like "revolution" and "new era" are used in the titles. Also see a couple of very recent articles[4,5].)
These advances have made it possible to obtain 3D structures of biological macromolecules in 3-5 Å range. For example, this is the structure of the yeast mitochondrial large ribosome subunit solved to the resolution of 3.2 Å by Ramakrishnan's group in collaboration with Sjors Scheres' group[6].
Since I attended Ramakrishnan's seminar, there have been a number of papers by various groups which reported high resolution structures using cryo-electron microscopy. For example, the following figure is the structure of E. coli ribosome by Fischer et al. that achieved the resolution of less than 3 Å[7].
The following is the structure of the human ribosome by Khatter et al.[8] with an average resolution of 3.6 Å, reaching 2.9 Å resolution at some places.
It's not just the ribosome. The following is the structure of 20S proteasome by Campbell et al. that was solved to 2.8 Å[9].
The following is the structure of anthrax protective antigen by Jiang et al. that was solved to 2.9 Å[10].
And then came this week the structure of β-galactosidase that achieved a resolution of 2.2 Å[11].
With this kind of resolution, you are able to see a very detailed structure.
I am ignorant about this field, but it seems that there have been tremendous improvements in the technique. This can have a huge impact on understanding the mechanisms of many biological macromolecules.
References
- Kühlbrandt, W. Biochemistry. The resolution revolution. Science 343, 1443-1444 (2014). [Pubmed] [Article]
- Kühlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014). [Pubmed] [Article]
- Bai, X. C., McMullan, G., & Scheres, S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 40, 49-57 (2015). [Pubmed] [Article]
- Cheng, Y., Grigorieff, N., Penczek, & P. A., Walz, T. A Primer to Single-Particle Cryo-Electron Microscopy. Cell 161, 438-449 (2015). [Pubmed] [Article]
- Cheng, Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 161, 450-457 (2015). [Pubmed] [Article]
- Amunts, A., Brown, A., Bai, X. C., Llácer, J. L., Hussain, T., Emsley, P., Long, F., Murshudov, G., Scheres, S. H., & Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485-1489 (2014). [Pubmed] [Article]
- Fischer, N., Neumann, P., Konevega, A. L., Bock, L. V., Ficner, R., Rodnina, M. V., & Stark, H. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520, 567-570 (2015). [Pubmed] [Article]
- Khatter, H., Myasnikov, A. G., Natchiar, S. K., & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015). [Pubmed] [Article]
- Campbell, M. G., Veesler, D., Cheng, A., Potter, C. S., & Carragher, B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife (2015). [Pubmed] [Article]
- Jiang, J., Pentelute, B. L., Collier, R. J., & Zhou, Z. H. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature (2015) [Epub ahead of print]. [Pubmed] [Article]
- Bartesaghi, A., Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L., & Subramaniam S. Science (2015) [Epub ahead of print]. [Pubmed] [Article]
Subscribe to:
Posts (Atom)