何ヶ月か前に、ヴェンキ・ラマクリシュナンのセミナーに出席する機会があった。ラマクリシュナンはリボソームの構造を解明した業績でノーベル賞を取った人だ。ノーベル賞を取るような科学者を生で見るのには感慨があるけれど、正直なところ、セミナーの内容はそれほど期待していたわけではなかった。彼の研究対象は僕自身の興味からは距離がある。細胞の中でタンパク質の工場の役割をするリボソームが重要なのは言うまでもないけれど、専門家でない人間に取ってリボソームの構造をいくつも見せられても得る物があるか疑問だった。それに彼のようにすでに功成り名を遂げた人物なら、その成功に満足して新しい成果を上げなくなっていてもおかしくはない。
予想は良い意味で裏切られた。ラマクリシュナンのノーベル賞の対象になった業績はX線結晶構造解析の手法で得られた物だけれど、驚いた事に、セミナーではクライオ電子顕微鏡を使った最近の研究の話をした。そもそも彼の事をX線結晶構造解析の専門家に分類するのが間違いなのだろう。彼はまず第一にリボソームに魅惑されていて―彼の熱意は言葉の端々に感じられた―リボソームの研究に役立つ手法ならなんでも取り入れるのだろう。彼は研究生活の最初からX線結晶構造解析をしていたわけではなかった。X線結晶構造解析もリボソームの研究のために学んだ手法に過ぎない。
なぜクライオ電子顕微鏡か。クライオ電子顕微鏡はX線結晶構造解析と比べて有利な点がいくつかある。まず第一に、サンプルを結晶化する必要がない。必要なサンプルの量も結晶化するために必要な量から比べればずっと少ない。X線結晶構造解析と違って、サンプルが均一でなくても研究できるし、いくつかの違ったコンフォメーションを解析できる可能性もある。とは言え、それで得られる構造が、有益な情報を得るのに十分な解像度がなければ、そういう利点も強みにはならない。実際、僕にとって、クライオ電子顕微鏡によって得られる構造というのは、細部がよくわからない、ぶよぶよとした塊のような物という印象があった。
僕には構造生物学は専門外で、この分野の進展に注意を払っていなかったので知らなかったのだけれど、クライオ電子顕微鏡によって達成できる解像度は近年著しく改善されてきたそうだ。ラマクリシュナンによると、この解像度の進歩はいくつかの技術的な発展によるものだそうだ。一つは、すぐれた検出器が開発されたこと。その他は、データを処理して構造を再構成するための手法の進展で、具体的には、ベイズ統計の導入や、電子ビームによって分子が動くことの影響を補正する事などだ。これらの進展は、最近のいくつかの概説にまとめられている。(例えば、 [1]、[2]、[3]。タイトルに「革命」とか「新時代」といった言葉が使われている事に注目。さらに、もっと最近の概説も参照[4,5]。)
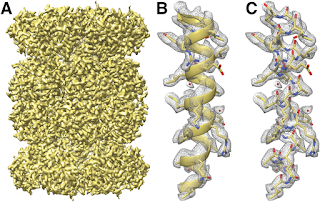
こういった進展によって、3-5オングストロム程度の解像度の生体分子の3次元構造を得る事が可能になった。例えば、以下に示すのはラマクリシュナンのグループがSjors Scheresのグループとの共同研究で得た酵母のミトコンドリアのリボソームの大サブユニットの構造[6]。
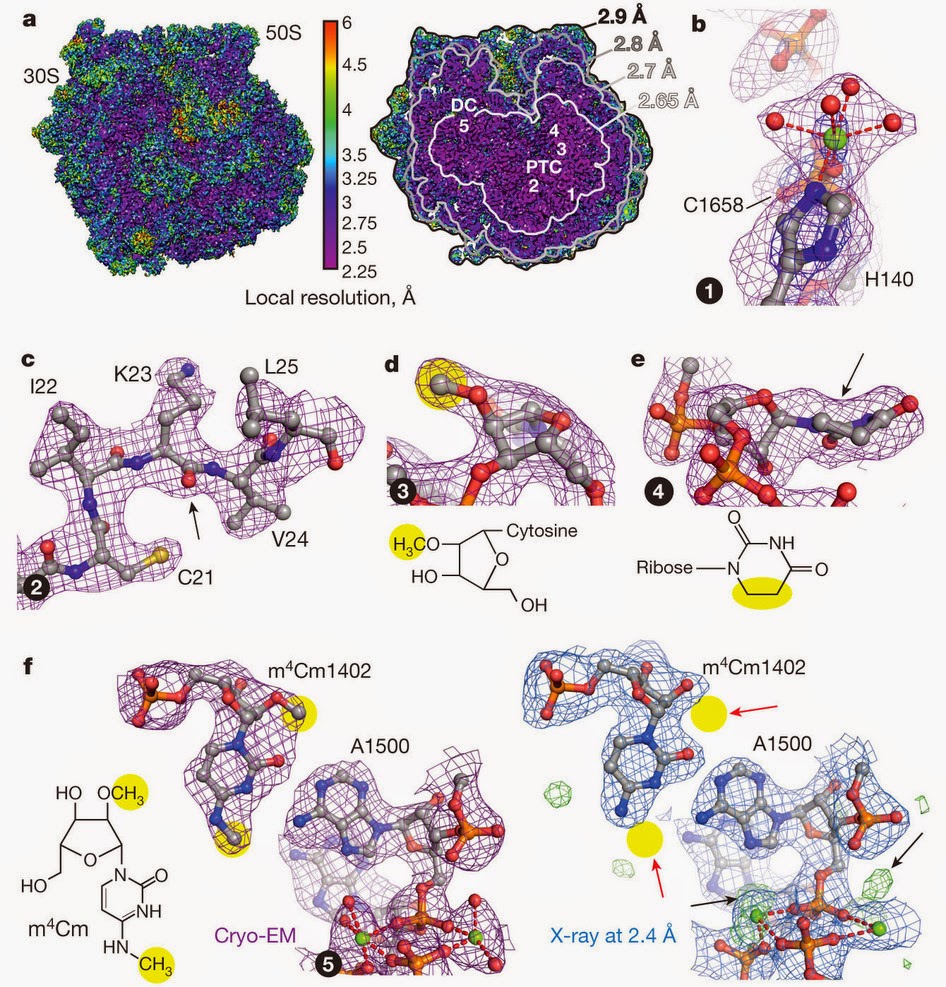
ラマクリシュナンのセミナーの後も、クライオ電子顕微鏡を使った高解像度の構造の論文をいろいろなグループが次々に発表している。例えば、次の図はFischerらによって発表された大腸菌のリボソームの構造で、解像度は3オングストローム未満を達成している[7]。
次の図はKhatterらによるヒトのリボソームの構造で[8]、解像度は平均で3.6オングストローム、部分によっては2.9オングストロームだ。
リボソームばかりではない。次の図はCampbellらによる20Sプロテアソームの構造で[9]、解像度は2.8オングストローム。
次の図は、Jiangらによる炭疽菌の防御抗原の構造で[10]、解像度は2.9オングストロームだ。
そして今週には解像度が2.2オングストロームのβガラクトシダーゼの構造が発表された[11]。
ここまで解像度が高いと、かなり細かな構造まで見える。
この分野については無知だけれど、ものすごい進歩が起きたみたいだ。生体分子のメカニズムの理解に大きなインパクトがありそうだ。
参考文献
- Kühlbrandt, W. Biochemistry. The resolution revolution. Science 343, 1443-1444 (2014). [Pubmed] [Article]
- Kühlbrandt, W. Cryo-EM enters a new era. eLife 3, e03678 (2014). [Pubmed] [Article]
- Bai, X. C., McMullan, G., & Scheres, S.H. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci. 40, 49-57 (2015). [Pubmed] [Article]
- Cheng, Y., Grigorieff, N., Penczek, & P. A., Walz, T. A Primer to Single-Particle Cryo-Electron Microscopy. Cell 161, 438-449 (2015). [Pubmed] [Article]
- Cheng, Y. Single-Particle Cryo-EM at Crystallographic Resolution. Cell 161, 450-457 (2015). [Pubmed] [Article]
- Amunts, A., Brown, A., Bai, X. C., Llácer, J. L., Hussain, T., Emsley, P., Long, F., Murshudov, G., Scheres, S. H., & Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 343, 1485-1489 (2014). [Pubmed] [Article]
- Fischer, N., Neumann, P., Konevega, A. L., Bock, L. V., Ficner, R., Rodnina, M. V., & Stark, H. Structure of the E. coli ribosome–EF-Tu complex at <3 Å resolution by Cs-corrected cryo-EM. Nature 520, 567-570 (2015). [Pubmed] [Article]
- Khatter, H., Myasnikov, A. G., Natchiar, S. K., & Klaholz, B. P. Structure of the human 80S ribosome. Nature 520, 640–645 (2015). [Pubmed] [Article]
- Campbell, M. G., Veesler, D., Cheng, A., Potter, C. S., & Carragher, B. 2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy. Elife (2015). [Pubmed] [Article]
- Jiang, J., Pentelute, B. L., Collier, R. J., & Zhou, Z. H. Atomic structure of anthrax protective antigen pore elucidates toxin translocation. Nature (2015) [Epub ahead of print]. [Pubmed] [Article]
- Bartesaghi, A., Merk, A., Banerjee, S., Matthies, D., Wu, X., Milne, J. L., & Subramaniam S. Science (2015) [Epub ahead of print]. [Pubmed] [Article]